Teaching
Thesis Internships availability
Thesis Internships for Bachelor and Master Degree students in Genomics, Biotechnology and Biology are available in the MEM-Lab.
Contact us for further information!

Prokaryotic Genomes (Bachelor Degree Programme in Genomics) - M. Cappelletti + M. Oggioni
Learning outcomes
By the end of the module the student has knowledge of the main approaches for genomic data analysis and annotation in prokaryotes, with an emphasis on the use of next generation sequencing for the functional and structural analysis of genomes. In particular the student is able to: understand the structure of genetic variability and its phenotypic effects, browse prokaryotic genomes, apply methods for genomic data analysis, and correctly interpret and plan genomic studies in bacteria.
Course contents
The Prokaryotic Genomes course (Module1+Module2) will focus on the following topics:
- Introduction to Prokaryotes: the prokaryotic cell, size, shape and arrangement of bacterial cells, structures internal and external to the cell wall, microbial growth
- Diversity of Prokaryotic World: bacterial and archeal taxonomy and phylogeny, the tree of life, molecular chronometers and phylogenetic tree, ecological niches, diversity based on nutrition, metabolism and physiology
- Genome sequencing: Genome sequencing technologies, Genome Reconstruction/assembly, Genome Annotation
- Prokaryotic genomes and plasticity: genome structure and partitioning, bacterial plasmids and transposable elements, Genomic Islands, core genomes VS accessory genomes, bacterial conjugation, transduction and transformation
- Comparative genomics of Prokaryotes and Microbial Genome Databases
- Mutagenesis as a Genomic Tool for Studying Gene Function: types of mutation, Transposon Mutagenesis and Targeted Mutagenesis
- Functional Genomics: Transcriptomics (RNA-seq and Microarray), Proteomics, Metabolomics, etc.
Systems and Synthetic Microbiology (Genomics Degree Programme) - M. Cappelletti
Learning outcomes
By the end of the course, the student has knowledge on the integrative approaches and strategies to study a microorganism as a complex system and to discover and model its properties in the context of synthetic biology, gene regulation, and microbial genome engineering across multiple scales. In particular, the course provides the student with systems and synthetic biology understanding of microorganisms and microbial ecosystems with industrial and environmental interest for biotechnological applications.
Course contents
Introduction to the basic concepts in the field of systems and synthetic microbiology; genome-wide engineering approaches and targeted genome editing methods for synthetic microbiology purposes; the idea of networks and their representation as graphs; the structure, dynamics, and evolution of metabolic networks; the building of biological parts and their function on the microbial systems level; the synthetic biology as a tool to build biological systems; synthetic biology tools for microbial genetic and metabolic engineering (e.g. BioBrick, pSEVA); genetic and metabolic circuits in microbial systems; design, construction and expression of synthetic pathways in microorganisms; synthetic and minimal genomes; systemic disciplines applied to microbial biotechnology (several case studies will be the subject of students analysis)
Microbial Biotechnologies (Master's Degree Programme in Molecular and industrial biotechnology) - S. Fedi
Learning outcomes
At the end of the course the student will have the basic knowledge on a few topics of industrial and environmental biotechnologies. In particular the student will know the molecular and metabolic features of those microorganisms used for i) biodegradtion of contaminants, ii) production of alternative energy sources as biogas, iii) enzyme production, and iv) development of commercial products with a high social impact.
Course contents
- Course organization along with details on the assessment methods. Introduction to the various topics.
- Microbial degradation of linear and branched hydrocarbons. Degradation of aromatics. Dehalogenation of haloaromatics. Genetic regulation of the catabolic pathways for the organic contaminants degradation. Catabolic pathways' evolution.
- Microorganism used in agriculture. Production of insecticides of microbial origin. Microbial production of antibiotics.
- Biotransformation of inorganics and their potential use in nanotechnologies
- Bacterial interaction with bacterial cells. Mechanism of oxidation/reduction of inorganics to be used for bacterial growth. Nanowires: generalities and applications.
- Bacterial cells and generation of biogas and electricity
- Interaction of bacterial cells with anodes and cathodes for the production of electricity (MFC) and/or valuable commercial chemicals. Bacterial production of biogas: generalities and microroganisms used for CH4 and H2 production in anerobiosis and phototrophy.
Laboratory
The student will acquire:
1) basic methods for the isolation and characterization of environmental samples with specific physiological features.
2) a few molecular methods to characterize complex microbial communities aiming to search for specific functional genes involved in biotech applications
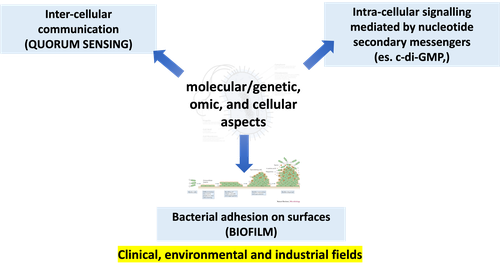
MICROBIAL SENSING, SIGNALLING AND INTERACTIONS (Master's Degree Programme in Molecular and Cell Biology - M. Cappelletti)
Learning outcomes
By the end of the course, the student has molecular and "omic" knowledge on the impact of microbial activity and biodiversity on human health, human activities and the environment. In particular, the course provides the student with: - understanding on the mechanisms at the basis of the bacterial cell signaling and the interaction between microorganisms and microorganism/environment; - competence on methods of advanced genetic engineering, functional genomics and synthetic biology to study microorganisms with clinical, environmental and industrial interest.
Course contents
The course covers molecular, cellular and “-omics” aspects of the following topics (considering both theoretical and methodological points of view): 1) Cell-cell communication in bacteria (quorum sensing): basic principles and components of quorum sensing (QS); role of the QS in microbial pathogenicity, genome plasticity (horizontal gene transfer), stress response, and microbial interaction with the host; application of quorum sensing circuits in biotechnology and synthetic biology of single bacteria and microbial communities. 2) Microbial biofilms: distribution and diversity of biofilms; mechanisms of biofilm formation and persistence; microbial metabolism and physiology in biofilm; role of QS in biofilm formation; biofilm resistance and tolerance; in vitro systems to grow and study the microbial biofilm; the role/importance of biofilms in medical and industrial fields. 3) Bacterial second messengers: molecular mechanisms of the nucleotide second messenger (NSM)-based intracellular signalling in bacteria; the different components involved in the NSM-based signalling; essential and emerging roles of NSMs in bacterial sensing and cellular response, biofilm formation, and microbial interactions. 4) Signalling and interactions within microbial communities: “-omics” to study microbial communities and microbial interactions; designing and construction of synthetic microbial communities for the application in medical, industrial, and environmental fields.
Readings/Bibliography
Textbook indications are given during the lectures. Scientific papers and Reviews can be obtained on-line. Alternatively, the material will be given to students as printed papers.
Teaching methods
Lectures, group works, and student presentations
https://www.unibo.it/en/study/phd-professional-masters-specialisation-schools-and-other-programmes/course-unit-catalogue/course-unit/2024/492228
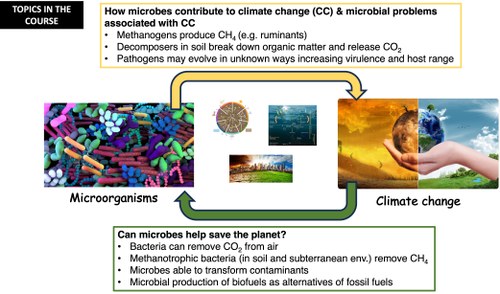
CLIMATE CHANGE MICROBIOLOGY IN HUMAN HEALTH (Bachelor Degree interuniversity programme in Biology of Human and Environmental Health)
Learning outcomes
Module 1 (Prof. S. Fedi)
The vast majority of terrestrial biodiversity is microbial in origin. Microorganisms are directly involved in the ecosystem processes on which all life depends. They interact intimately with other organisms, yet we know far less about their biodiversity than that of plants and animals. Our lack of understanding of the interactions and feedbacks of microbial biodiversity is worrying given the high rates of environmental change the Earth is currently experiencing, including human-caused global climate change. The Course will mainly analyse the impact that climate change has and may have on the one hand on the different geochemical cycles (carbon Nitrogen, sulphur, phosphorus) and on the other hand on the spread and alteration of microbial diseases in the different continents depending on climate change.
Module 2 (Prof. M. Cappelletti)
Lectures will present laboratory activities and provide basic knowledge of methods (molecular and traditional) for analysing the genetics and functions of microorganisms in environmental samples.
Course contents
Module 1
Influence of microorganisms on climate. Impact of climate change on microorganisms and geochemical cycles (carbon, nitrogen sulphur, phosphorus). Importance of microorganisms in the transformation processes of compounds that are toxic to humans and what influence climate change has on them; Influence of climate change on the spread of major diseases of microbial origin (bacteria, fungi and viruses). How climate hazards, e.g. changes in temperature and the availability of water and food affect the development of infectious diseases in humans. Climate change-dependent increase in the spread and even treatment resistance of the main pathogens (e.g. Staphylococcus aureus, Escherichia coli, Klebsiella pneumoniae, Streptococcus pneumoniae and non-typhoidal Salmonella).
Module 2
During the laboratory activities, students will apply techniques for bacterial isolation and enrichment of specific microbial groups (e.g., nitrogen-fixers, Actinobacteria) from different environmental matrices (e.g., soil, plants). Metabolic/enzymatic assays of the isolated bacterial strains will be performed in order to identify bacteria producing antibiotics and specific enzyme activities (e.. amylase and urease) involved in biogeochemical cycles. Finally, analysis of the soil microbiota will be performed by DNA extraction and amplification of the 16S rRNA gene that will be cloned to construct libraries in E.coli. The clone libraries will be then analyzed by molecular methods to determine the different bacterial groups present in the soil sample.