Research activity
Genome instability mechanisms and their connections to immunological pathways.
Regulation of genomic functions is a complex process that involves several protein factors and nucleic acid structures. Particularly in eukaryotes, our understanding of functional roles of protein factors is much more advanced than the role of genomic DNA itself and RNA molecules. However, in the last decade, thanks mainly to the development of parallel sequencing technologies (known as Next Generation Sequencing, NGS), it has become possible to investigate how particular secondary structures of nucleic acids can affect fundamental genomic functions such as gene expression, DNA replication, genome recombination and stability. Non-B DNA structures have been established to play major roles in cell transformation and neuron degeneration as they markedly influence expression profiles and genome stability.
In this context, our research focuses on how non-B DNA structures affect chromatin organization, genome functions and stability, on one side, and how they are regulated by DNA topoisomerases, on the other. We are particularly interested in R loops and G-quadruplex (G4). R-loops are formed during transcription upon re-annealing of the nascent RNA to the DNA template strand, forming an RNA:DNA hybrid and forcing the non-template strand to loop out in a single-stranded status (see Figure 1). G4s are instead generated by single-stranded guanine-rich DNA sequences that fold into stable four-stranded structures wherein three sets of four guanines are arranged in three stacking planar quartets (see Figure 2).
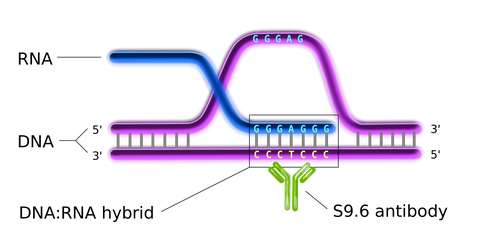
Figure 1. The general structure of an R loop (Wikimedia Commons).
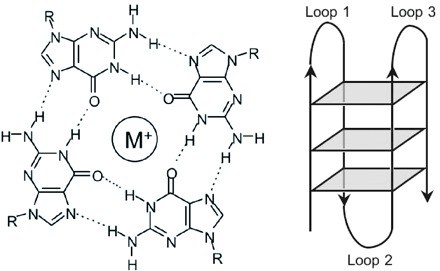
Figure 2. The general structure of G-quadruplex (G4). Note that G4 structures are very polymorphic (Wikimedia Commons).
NGS techniques have allowed the mapping of non-B structures along the genome. The mapping data show that R loops and G4s are prevalent in mammalian genomes, where they form dynamically over conserved regions. In particular, R-loops have been linked to genomic instability by causing interference between the replication and transcription machineries. R loops and G4s are DNA structures particularly sensitive to DNA supercoilings. Negative DNA supercoils generated behind an elongating RNA polymerase are thought to facilitate R-loop formation by inducing an underwound DNA duplex favorable to re-annealing of the nascent transcript to its template. Interestingly, huma genomic maps showed that active promoters are free of nucleosomes, have more negative supercoils than other genomic sites, and are regions where R loops and G4s can form more easily.
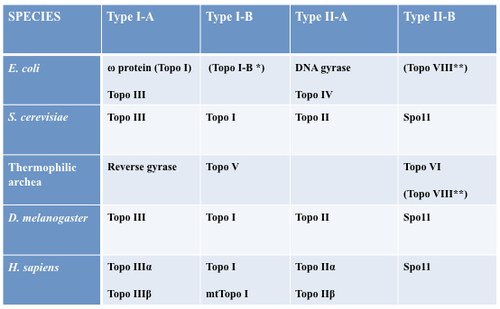
Figure 3. DNA topoisomerase family (to read more, Capranico G, Marinello J, Chillemi G., Type I DNA topoisomerase, J Med Chem. 2017 Mar 23;60(6):2169-2192).
Among several DNA topoisomerases present in a mammalian cell (see Figure 3), DNA Topoisomerase I is a main cellular factor controlling topological homeostasis of the genome. Thus, we have recently studied the role of Topoisomerase I in R loop formation in human cells by siRNA gene silencing and NGS technology. The findings were somewhat surprising as they demonstrate that Topoisomerase I can indeed regulate an R-loop, but it can either favor or dis-favor its formation depending on chromatin context. The paper has been published in Genome Biology in a close collaboration with Fred Chedin who hosted Stefano Manzo, a former student of our PhD program in Cellular and molecular biology, for a year at UCS in Davis, CA, USA.
We recently investigated the role of anticancer TOP1 poisons (CPT and LMP776) in micronuclei generation with a mechanism involving R-loop accumulation, leading to activation of the cGAS-STING pathway and immune gene expression in HeLa cancer cells. In addition, we show that Top1 poisons can activate both IRF3- and NF-kB-dependent genes, however, they cannot in SCLC cancer cells as the cGAS-STING pathway is markedly impaired likely due to several mechanisms including a drastic reduction of STING and/or cGAS expression. These data were published in British Journal of Cancer in 2022.
As a number of structural features of the genome, such as G-richness, GC skew and negative supercoils, can favor the formation of both R-loops and G4, we wondered whether chemical stabilization of one structure (G4) can trigger the formation of the other (R-loop) in living cells. Thus, A new PhD student in the lab, Alessio De Magis, together with Stefano Manzo, performed several experiments to investigate the effects of G4 ligands on R-loop formation and genome integrity in human cancer cells using R-loop immunoprecipitation methods (called DRIP) and NGS technology. By studying three structurally-unrelated G4 ligands and one inactive analog, we established that G4 ligands induce an immediate increase of nuclear R loops as G4s and R-loops can cooperate and stabilize each other during transcription, forming a G-loop (Figure 4). Moreover, the data show that DNA damage induced by G4 ligands are mediated by R-loops. We also discovered that G4 ligands cause the generation of micronuclei at later times in an R loop-mediated manner, particularly in BRCA2-depleted cancer cells.This work has been published in the Proceedings of the National Academy of Sciences USA. We have thus establish a mechanistic role for R loops in mediating the cellular effects of G4 ligands, which have been reviewed in Nucleic Acids Research recently.
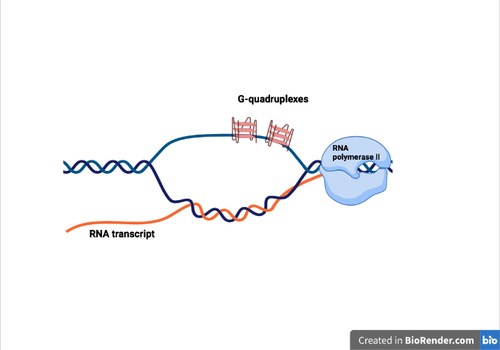
Figure 4. A G-loop structure can form behind an elongating RNA polymerase.
The previuous data open innovative lines of investigation to develop G4 ligands as new types of anticancer agents. More recently, Giulia Miglietta and Marco Russo (two post-docs in the lab) investigate the consenquences of micronuclei produced by G4 ligands in human breast cancer cells. They studied two different G4 binders showing that they induce an increase of micronuclei triggering the activation of the cytoplasmic DNA sensor, cGAS, and the STING factor in human and murine cancer cells (see Figure 5). Ligand activity can then lead to Interferon B (IFNB) and other IFNB-responsive gene activation. Interestingly, specific gene expression patterns medi-ated by G4 binders in cancer cells correlate with immunological hot features and better survival in human TCGA (The Cancer Genome Atlas) breast cancers. The findings (published in Nucleic Acids Research) open to the development of cytostatic G4 binders as effective immunomodulators for combination immunotherapies in unresponsive tumors.
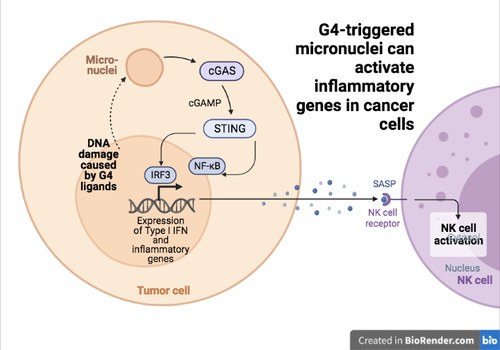
Figure 5. The cGAS-STING pathway is activated by micronuclei induced G4 ligands of Topoisomerase I poisons in human cancer cells.
In summary, our research goal is to establish molecular and epigenetic mechanisms dependent on non-B DNA structures of transcription regulation and genome instability. As several enzymes and other factors recognize these structures and regulate their stability and folding, mutations of these enzymes can affect diseases progression and therapeutic intervention. Thus, the discovery of regulation mechanisms involving R loops and G4s can reveal unexpected opportunities to develop strategies for personalized medicine in oncology, immunological disorders and neurodegenerative diseases.
Research activity in the lab is supported by funds from Associazione Italiana per la Ricerca sul Cancro (AIRC), Fondazione Italiana per la Ricerca sul Cancro (FIRC), PRIN program of the Ministero dell'Università e della Ricerca (MUR), Regione Emilia-Romagna and University of Bologna.
HIGHLIGHTS
-
Miglietta G., M. Russo, R.C. Duardo and G. Capranico, G-quadruplex binders as cytostatic modulators of innate immune genes in cancer cells. Nucleic Acids Research, 2021, 49, 6673–6686.
Here, we present evidence that G4-binder induced micronuclei activate immune gene expression in human cancer cells.
-
Miglietta G, Russo M and Capranico G, G-quadruplex–R-loop interactions and the mechanism of anticancer G-quadruplex binders. Nucleic Acids Research, 2020, 48, 11942–11957.
The review proposes that the induction of R-loops is a main mechanism of action of G4 binders with therapeutic activity.
-
De Magis A, Manzo SG, Russo M, Marinello J, Morigi R, Sordet O, and Capranico G, DNA damage and genome instability by G-quadruplex ligands are mediated by R loops in human cancer cells. PNAS, 2019, 116, 816-825.
The data show that G-loops (a new structure made of G-quadruplexes in the displaced strand of an R-loop) are induced by G4 binders in living human cancer cells.
-
Manzo SG, Hartono SR, Sanz LA, Marinello J, De Biasi S, Cossarizza A, Capranico* G and Chedin* F, DNA Topoisomerase I differentially modulates R-loops across the human genome. Genome Biology (2018) 19:100
The paper defines the genome-wide role of Top1 in regulating R loops in human cells.