SARS-CoV-2
"Genomic snapshot of the SARS-CoV-2 sequencing effort in the Emilia-Romagna region of Italy", by Daniele Mercatelli, Simone Di Giacomo and Federico M. Giorgi Department of Pharmacy and Biotechnology, University of Bologna, Italy (2022)
Abstract
Genomic surveillance is paramount in tracking and understanding the evolution of COVID-19 in specific geographic regions, in order to predispose appropriate clinical and social responses. In this study, thanks to the data collection and effort of all the centers involved, we could perform the first multi-sample meta-analysis of SARS-CoV-2 genomic sequences from the Emilia-Romagna region, analyzing 5,646 samples. Our analysis, mainly covering the 13-months of January 2021-January 2022, shows the prevalence of Delta variant SARS-CoV-2 samples in the region during 2021, and a recent rapid growth of Omicron BA.1 variant. Furthermore, we highlight the presence of at least four distinct cases of Omicron BA.2, characterized by several specific amino acid features, especially in the viral Spike protein.
Keywords
SARS-CoV-2; COVID-19; Omicron; Emilia-Romagna; Italy; Genomics; BA.1; BA.2; Genomic surveillance
Introduction
More than two years have passed since the discovery of a novel beta-coronavirus in the province of Wuhan, China [1]. First named 2019-nCov [2], and then SARS-CoV-2 [3] by the World Health Organization (WHO), the virus causes COVID-19, a contagious respiratory disease which quickly evolved to a worldwide concern. COVID-19 has become the first pandemic in human history to be vastly investigated and monitored by genomic sequencing [4], allowing to track the genomic evolution of SARS-CoV-2 alongside its geographic, temporal, and clinical expansion.
According to initial estimates [5], the 29903 nucleotide long single-stranded RNA genome of SARS-CoV-2 has maintained a mutation rate similar to other coronaviruses, of approximately 1.5 x 10−3 substitutions per site per year [6,7]. This has generated roughly 30 new mutations/year, which combined to give rise to several novel variants [8]; a standardized and evolutionarily rational nomenclature for all variants and lineages has been established by the Pangolin project [9]. Amongst all variants, Variants of Concern (VOCs) have been named, according to the WHO, with progressive Greek alphabet ids. The first was Alpha (Pangolin B.1.1.7 lineage), also named "British variant" due to its discovery by the enormous sequencing effort of the United Kingdom [10]. Alpha contained 22 aminoacidic (aa) mutations compared to the original Wuhan sequence, spread across four viral proteins, including the Spike Glycoprotein, responsible for the initial ligand binding with the human cell host receptors ACE2 and TMPRSS2 [11]. Following Alpha appearance in late 2020, the Beta variant (B.1.351) carried 18 aa mutations and has also risen to the status of VOC [8], but both have rapidly recessed during 2021, to leave way to the highly spreading Delta variant (B.1.617.2), which, with its 29 aa mutations, quickly became the dominant lineage globally [12]. In late 2021/early 2022, global concern has instead focused on a yet new variant, named Omicron (B.1.1.529), which showed higher transmissibility and higher immunoevasion potential when compared to previous VOCs [13]; the first detected Omicron virus showed 49 mutations compared to the original strain, a large number of which (30) are located on the Spike protein [14].
In this rapidly changing context of SARS-CoV-2 evolutionary steps, genomic surveillance is of paramount importance, as it can discover new mutations and variants, as well as monitoring the geographic and temporal progression of COVID-19 strains, each of which is characterized by different clinical features [15]. While global genomic aggregation studies can provide a unique insight into understanding SARS-CoV-2 evolution [16], local studies have a unique usefulness, as they can provide a snapshot of variant status in a geographically and socially uniform context [17,18].
In the current communication, we will provide an overview of SARS-CoV-2 variant prevalence in the Emilia-Romagna region, by aggregating all genomic data derived from local sequencing centers. Located between the 44th and 45th parallel north, between Po Valley and the Apennine Mountain range, Emilia-Romagna is the second richest region of Italy according to GDP per capita [19] and deploys one of the most efficient healthcare systems of Europe [20]. Our analysis will investigate the genomic status of SARS-CoV-2 in this region at the time of writing (February 2022).
Materials and Methods
Regional Vaccination and Case data were obtained from the Italian Government, Presidency of the Council of Ministers, COVID-19 database (https://www.governo.it/it/cscovid19/report-vaccini/). Raw full-length (>29000 nucleotides) SARS-CoV-2 sequences were obtained, in FASTA format, from the GISAID database [21], from contributing centers indicated in Supplementary File S1.
Sequences were compared to the Wuhan SARS-CoV-2 genome, NCBI id NC_045512.2, using a previously published pipeline [22]. Detected mutations allowed then the associations of each sample to the most likely variant using the Pangolin COVID-19 Lineage Assigner (https://pangolin.cog-uk.io/)[9].
All statistical and graphical analyses were performed with R, version 4.1.1. The structure between SARS-CoV-2 Omicron Spike protein and human ACE2 was derived from the Cryo-EM experiment of Hong and colleagues [23], uploaded on the Protein Data Bank under entry 7WK6 [24].
Results
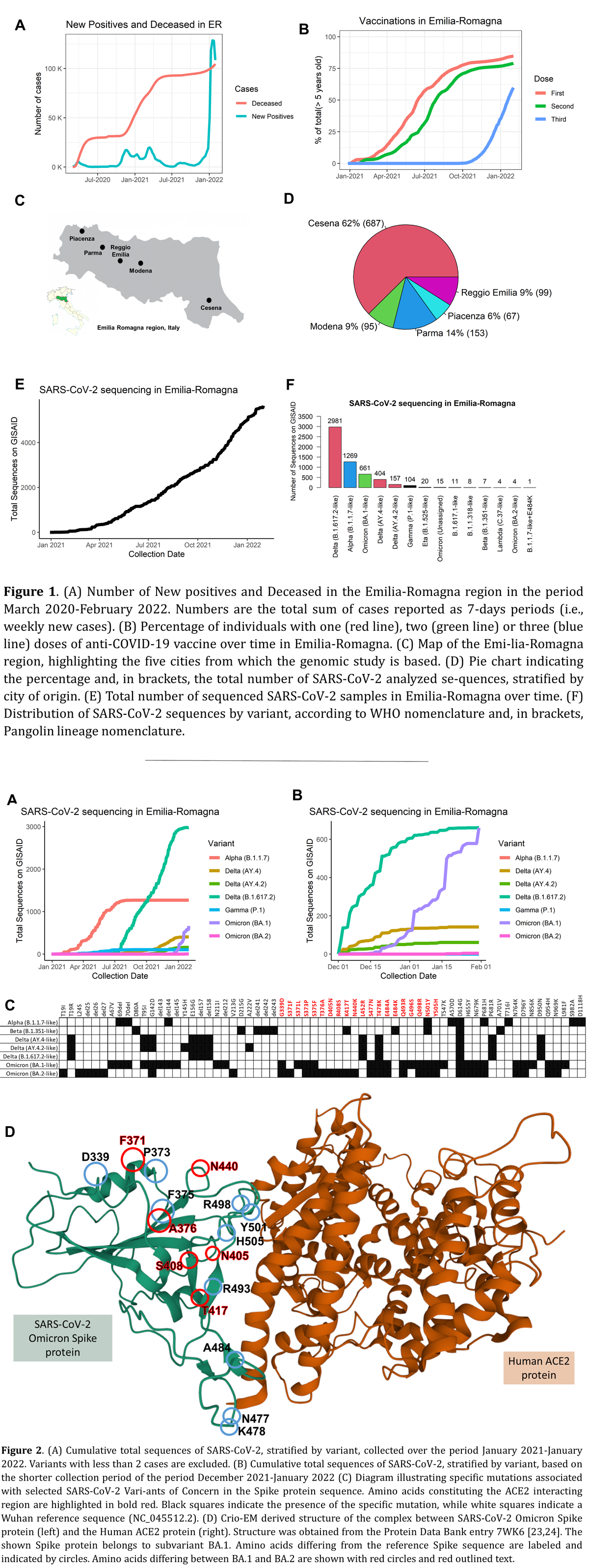
Alongside the rest of the World, Emilia-Romagna suffered multiple waves of COVID-19 starting early 2020 (Figure 1 A), with more than 80% of its population having now received at least one vaccine dose (Figure 1 B). In this epidemiological context, genomic sequencing of SARS-CoV-2 has been undertaken and results have been publicly shared on the GISAID database [21] on samples from 5 cities: Cesena (96K inhabitants), Modena (185K inhabitants), Parma (194K inhabitants), Piacenza (102K inhabitants), and Reggio Emilia (171K inhabitants) (Figure 1 C). A total of 5646 full length SARS-CoV-2 genome sequences were obtained. A small portion of these sequences (1101) could be associated with a specific city of origin: specifically, more than half (62%) derive from the Romagna AUSL in the territory of Cesena (in the east of the Emilia-Romagna region) and the rest (38%) from centers in the west (Figure 1 D).
The sequencing effort in the region has effectively started in January 2021, with samples collected largely since then, and covering therefore roughly 13 months at the time of writing (Figure 1 E). In this time window, we could detect a prevalence of all major SARS-CoV-2 variants of concern Alpha, Beta, Gamma, Delta, and Omicron (Figure 1 F and 2 A). Sequences associated to the Omicron variant sequences have reached in number and frequency the previously dominant Delta variant sequences during the second half of January 2022 (Figure 2 B), with negligible numbers of Alpha and Gamma variant sequences (Figure 2 B).
Alongside the clear presence of the major type of SARS-CoV-2 Omicron virus (BA.1) in 661 distinct Emilia-Romagna cases, we could also detect 4 sequences that can clearly be associated to a novel Omicron subvariant, BA.2. Recently, Omicron has been separated in 3 subvariants, BA.1, BA.2 and BA.3., sharing a core of more than 40 mutations. BA.1 was the first Omicron identified, followed by BA.2, which has recently achieved predominance in Denmark and other countries [25]. BA.3 is much less common worldwide and was not detected in our study.
While characterized by large similarity, BA.1 and BA.2 differ in more than 10 aminoacid loci in the Spike protein alone (Figure 2 C). Several amino acids differing between BA.1 and BA.2 are located on the interaction surface with the human protein ACE2 (Figure 2 D), such as (using BA.2 sequence status nomenclature): F371, A376, N405, S408, and N440.
Early evidence showed that Omicron is 4 times more infectious than the Wuhan original virus, and 2 times more than the Delta variant [26]. Amongst Omicron subvariants, a Danish study currently in preprint suggests a higher transmissibility for the BA.2 variant compared to the BA.1 [25]. However, there currently is no published evidence, to our knowledge, of clinical differences between Omicron BA.1 and BA.2 subvariants.
Discussion
The discovery of the Omicron subvariant of SARS-CoV-2 in Emilia-Romagna, with two reported cases of the BA.2 subvariant, testifies the need for continued genomic surveillance and the importance of sequencing efforts in monitoring and managing the COVID-19 pandemic. Only by aggregating multicenter efforts and aggregating large number of sequencing data, even in geographically circumscribed regions, it will be possible to monitor the waves of SARS-CoV-2 variants over time with reliable evidence (such as shown in Figure 1 F). Fortunately, current vaccines have been shown to provide protection also against all current major VOCs [27]. Still, localized snapshots of genomic frequencies of present and future COVID-19 variants can be translated into clinical response readiness for peculiarities of SARS-CoV-2 infection, such as pharmacological response, efficacy of vaccines and overall outcome [25].
Author Contributions: Conceptualization, DM, SdG, FMG; methodology, DM, FMG; software, FMG; validation, SdG; formal analysis, DM, SdG, FMG; investigation, DM, SdG, FMG; resources, FMG; data curation, DM, FMG; writing—original draft preparation, FMG; writing—review and editing, DM, SdG, FMG; visualization, DM, FMG; supervision, DM, SdG, FMG; project admin-istration, DM, SdG, FMG; funding acquisition, FMG. All authors have read and agreed to the published version of the manuscript.
Funding: This work was supported by the Fondazione CARISBO under the Bando Ricerca Medica e Alta Tecnologia 2021 (project 2021.0167).
Data Availability Statement: All data are available at the GISAID portal (https://www.epicov.org/).
Acknowledgments: The authors are very grateful to the GISAID Initiative and all its data con-tributors, that is, the authors from the originating laboratories responsible for obtaining the specimens and the Submitting laboratories where genetic sequence data were generated and shared via the GISAID Initiative, on which this study is based. Specifically, for this study we thank all which contributed to making SARS-CoV-2 genomic sequences to the GISAID portal for the different Emilia-Romagna cities: 1) Cesena: the U.O. Microbiologia Laboratorio Unico Centro Servizi - AUSL della Romagna; 2) Reggio Emilia: Arcispedale Santa Maria Nuova Autoimmunità Allergologia e Biotecnologie Innovative; 3) Modena: the Azienda Ospedaliero/Universitaria of Modena Policlinico - Virologia e Microbiologia Molecolare; 4) Piacenza: the Azienda Sanitaria Locale of Piacenza - Presidio Ospedaliero – Microbiology Lab, the IGA Technology Services Srl of Udine; 5) Parma: the University of Parma - Laboratorio di Igiene e Sanita Pubblica.
The authors wish to thank Dr. Lucia Ferroni and all the Department of Pharmacy and Biotechnology at the University of Bologna for the administrative support.
Conflicts of Interest: The authors declare no conflict of interest.
References
1. Du Toit A. Outbreak of a novel coronavirus. Nat Rev Microbiol 2020; 18:123–123
2. Gralinski LE, Menachery VD. Return of the Coronavirus: 2019-nCoV. Viruses 2020; 12:135
3. Kim D, Lee J-Y, Yang J-S, et al. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020; 181:914-921.e10
4. Oude Munnink BB, Worp N, Nieuwenhuijse DF, et al. The next phase of SARS-CoV-2 surveillance: real-time molecular epidemiology. Nat Med 2021; 27:1518–1524
5. Dearlove B, Lewitus E, Bai H, et al. A SARS-CoV-2 vaccine candidate would likely match all currently circulating variants. PNAS 2020; 117:23652–23662
6. Chiara M, Horner DS, Gissi C, et al. Comparative Genomics Reveals Early Emergence and Biased Spatiotemporal Distribution of SARS-CoV-2. Molecular Biology and Evolution 2021; 38:2547–2565
7. Jha N, Hall D, Kanakan A, et al. Geographical Landscape and Transmission Dynamics of SARS-CoV-2 Variants Across India: A Longitudinal Perspective. Frontiers in Genetics 2021; 12:
8. Tao K, Tzou PL, Nouhin J, et al. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat Rev Genet 2021; 22:757–773
9. O’Toole Á, Scher E, Underwood A, et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evolution 2021; 7:veab064
10. Fort H. A very simple model to account for the rapid rise of the alpha variant of SARS-CoV-2 in several countries and the world. Virus Research 2021; 304:198531
11. Guzzi PH, Mercatelli D, Ceraolo C, et al. Master Regulator Analysis of the SARS-CoV-2/Human Interactome. Journal of Clinical Medicine 2020; 9:982
12. Torjesen I. Covid-19: Delta variant is now UK’s most dominant strain and spreading through schools. BMJ 2021; 373:n1445
13. VanBlargan LA, Errico JM, Halfmann PJ, et al. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat Med 2022; 1–6
14. Mullen JL, Tsueng G, Latif A, et al. outbreak.info.
15. del Rio C, Omer SB, Malani PN. Winter of Omicron—The Evolving COVID-19 Pandemic. JAMA 2022; 327:319–320
16. Mercatelli D, Giorgi FM. Geographic and Genomic Distribution of SARS-CoV-2 Mutations. Frontiers in Microbiology 2020; 11:
17. López-Causapé C, Fraile-Ribot PA, Jiménez-Serrano S, et al. A Genomic Snapshot of the SARS-CoV-2 Pandemic in the Balearic Islands. Frontiers in Microbiology 2022; 12:
18. Gupta A, Sabarinathan R, Bala P, et al. A comprehensive profile of genomic variations in the SARS-CoV-2 isolates from the state of Telangana, India. J Gen Virol 2021; 102:001562
19. Mosconi F, D’Ingiullo D. Institutional quality and innovation: evidence from Emilia-Romagna. Economics of Innovation and New Technology 2021; 0:1–33
20. Meschi T, Rossi S, Volpi A, et al. Reorganization of a large academic hospital to face COVID‐19 outbreak: The model of Parma, Emilia‐Romagna region, Italy. Eur J Clin Invest 2020; 50:e13250
21. Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data – from vision to reality. Eurosurveillance 2017; 22:
22. Mercatelli D, Triboli L, Fornasari E, et al. Coronapp: A web application to annotate and monitor SARS-CoV-2 mutations. Journal of Medical Virology 2021; 93:3238–3245
23. Hong Q, Han W, Li J, et al. Molecular basis of SARS-CoV-2 Omicron variant receptor engagement and antibody evasion and neutralization. 2022; 2022.01.10.475532
24. Burley SK, Bhikadiya C, Bi C, et al. RCSB Protein Data Bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Research 2021; 49:D437–D451
25. Lyngse FP, Kirkeby CT, Denwood M, et al. Transmission of SARS-CoV-2 Omicron VOC subvariants BA.1 and BA.2: Evidence from Danish Households. 2022; 2022.01.28.22270044
26. Garcia-Beltran WF, St Denis KJ, Hoelzemer A, et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. medRxiv 2021; 2021.12.14.21267755
27. Pajon R, Doria-Rose NA, Shen X, et al. SARS-CoV-2 Omicron Variant Neutralization after mRNA-1273 Booster Vaccination. New England Journal of Medicine 2022; 0:null