
the COBRAMM GitLab wiki pages host the Installation Guide and the Manual, including a brief introduction to the QM/MM scheme and COBRAMM basic usage guide.

The tutorial cover the MM setup and the QM/MM calculations of a benzophenone molecule solvated in water, to compute absorption and emission properties with TD-CAM-B3LYP using Gaussian, and finally excited state molecular dynamics.

The tutorial introduces some COBRAMM functionalities with simple“static” and “dynamical” studies of the retinal photochemistry in protein environment, using the crystallographic data of Bovine Rhodopsin (Rh; PDB code: 1U19, chain A).
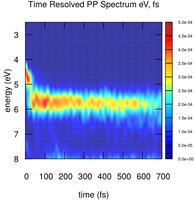
This tutorial is meant to introduce the COBRAMM utilities for the simulation of transient pump-probe spectroscopies (UV-Vis, Tr-PES).
Group leader of the Computational Photophysics and Photochemistry group
University of Bologna
Associate Professor at the Computational Photophysics and Photochemistry group
University of Bologna